Single Cell apoptosis¶
As decribed in Monier et al. 2015.
We start with a simple hexagonal apical mesh with cylindrical symmetry.
Imports¶
[1]:
import pandas as pd
import numpy as np
import json
import matplotlib.pylab as plt
%matplotlib inline
import ipywidgets as widgets
from IPython.display import display, Image
import tyssue
from tyssue import Sheet, History
from tyssue import config
from tyssue import SheetGeometry as geom
from tyssue.dynamics.sheet_vertex_model import SheetModel as basemodel
from tyssue.dynamics.apoptosis_model import SheetApoptosisModel as model
from tyssue.solvers.quasistatic import QSSolver
from tyssue.draw import sheet_view, create_gif, browse_history
from tyssue.io.hdf5 import load_datasets
print(tyssue.__version__)
0.7.1
Loading the datasets and specifications¶
Here it is a cylindrical hexagonal mesh.
[2]:
# Read pre-recorded datasets
h5store = 'data/small_hexagonal.hf5'
from tyssue.io.hdf5 import save_datasets, load_datasets
datasets = load_datasets(h5store,
data_names=['face', 'vert', 'edge'])
# Corresponding specifications
specs = config.geometry.cylindrical_sheet()
sheet = Sheet('emin', datasets, specs)
sheet.sanitize(trim_borders=True, order_edges=True)
geom.update_all(sheet)
# Model
nondim_specs = config.dynamics.quasistatic_sheet_spec()
dim_model_specs = model.dimensionalize(nondim_specs)
sheet.update_specs(dim_model_specs)
sheet.get_opposite()
live_edges = sheet.edge_df[sheet.edge_df['opposite']==-1].index
dead_src = sheet.edge_df.loc[live_edges, 'srce'].unique()
### Boundary conditions
sheet.vert_df.is_active = 1
sheet.vert_df.loc[dead_src, 'is_active'] = 0
sheet.edge_df['is_active'] = sheet.upcast_srce('is_active') * sheet.upcast_trgt('is_active')
First energy minimization¶
[3]:
min_settings = {
# "minimize":{
'options': {
'disp': False,
'ftol': 1e-6,
'gtol': 1e-5},
# }
}
solver = QSSolver()
res = solver.find_energy_min(sheet, geom, model, **min_settings)
print(res['success'])
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
True
Custom display function¶
[4]:
def leg_joint_view(sheet, coords=['z', 'x', 'y']):
geom.update_all(sheet)
x, y, z = coords
datasets = {}
datasets['face'] = sheet.face_df.sort_values(z)
datasets['vert'] = sheet.vert_df.sort_values(z)
edge_z = 0.5 * (sheet.upcast_srce(sheet.vert_df[z]) +
sheet.upcast_trgt(sheet.vert_df[z]))
datasets['edge'] = sheet.edge_df.copy()
datasets['edge'][z] = edge_z
datasets['edge'] = datasets['edge'].sort_values(z)
tmp_sheet = Sheet('tmp', datasets,
sheet.specs)
tmp_sheet.reset_index()
draw_specs = {
'vert': {
'visible': False
},
'edge': {
'color': tmp_sheet.edge_df[z],
#'zorder': depth.values
}
}
fig, ax = sheet_view(tmp_sheet, coords[:2], mode='2D', **draw_specs)
ax.set_xlim(-15, 15)
ax.set_ylim(-10, 10)
ax.set_facecolor('#404040')
ax.set_xticks([])
ax.set_yticks([])
fig.set_size_inches((10, 12))
return fig, ax
fig, ax = leg_joint_view(sheet)
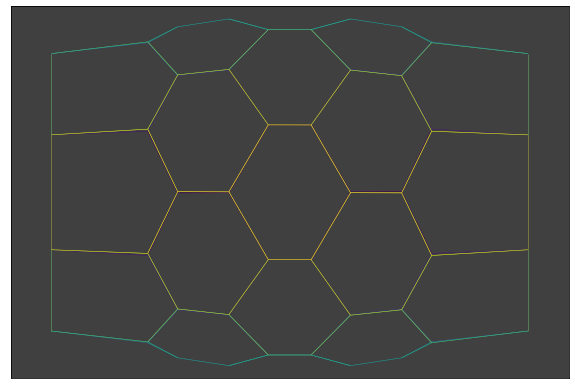
Choosing an apoptotic cell¶
[5]:
apoptotic_cell = 16
print('Apoptotic cell position:\n{}'.format(sheet.face_df.loc[apoptotic_cell, sheet.coords]))
apoptotic_edges = sheet.edge_df[sheet.edge_df['face'] == apoptotic_cell]
apoptotic_verts = apoptotic_edges['srce'].values
print("Indices of the apoptotic vertices: {}".format(apoptotic_verts))
Apoptotic cell position:
x -7.975392
y 3.332710
z -4.560289
Name: 16, dtype: float64
Indices of the apoptotic vertices: [33 42 34 41 43 44]
Cell behaviours, such as division or apoptosis, are defined as a series of unit changes in the tissue, such as the cell growth, or the abscission of the cell face once division is complete. For a given behavior, the next step often depends on the current state. This is managed by an EventManager object, which runs the behaviour.
[6]:
from tyssue.behaviors.sheet import apoptosis
from tyssue.behaviors import EventManager
manager = EventManager('face')
sheet.settings['apoptosis'] = {
'shrink_rate': 1.2,
'critical_area': 8.,
'radial_tension': 0.2,
'contractile_increase': 0.3,
'contract_span': 2
}
sheet.face_df['id'] = sheet.face_df.index.values
manager.append(apoptosis, face_id=apoptotic_cell, **sheet.settings['apoptosis'])
Here is the code for the apoptosis behavior:
def apoptosis(sheet, manager, **kwargs):
"""Apoptotic behavior
While the cell's apical area is bigger than a threshold, the
cell shrinks, and the contractility of its neighbors is increased.
once the critical area is reached, the cell is eliminated
from the apical surface through successive type 1 transition. Once
only three sides are left, the cell is eliminated from the tissue.
Parameters
----------
sheet : a :class:`Sheet` object
manager : a :class:`EventManager` object
face_id : int,
the id of the apoptotic cell
shrink_rate : float, default 0.1
the rate of reduction of the cell's prefered volume
e.g. the prefered volume is devided by a factor 1+shrink_rate
critical_area : area at which the face is eliminated from the sheet
radial_tension : amount of radial tension added at each contraction steps
contractile_increase : increase in contractility at the cell neighbors
contract_span : number of neighbors affected by the contracitity increase
geom : the geometry class used
"""
apoptosis_spec = default_apoptosis_spec
apoptosis_spec.update(**kwargs)
face = apoptosis_spec["face"]
if sheet.face_df.loc[face, "area"] > apoptosis_spec["critical_area"]:
# Shrink and pull
shrink(sheet, face, apoptosis_spec["shrink_rate"])
ab_pull(sheet, face, apoptosis_spec["radial_tension"])
# contract neighbors
neighbors = sheet.get_neighborhood(
face, apoptosis_spec["contract_span"]
).dropna()
neighbors["id"] = sheet.face_df.loc[neighbors.face, "id"].values
manager.extend(
[
(
contraction,
{
"face_id": neighbor["id"],
"contractile_increase": (
apoptosis_spec["contractile_increase"] / neighbor["order"],
),
},
)
for _, neighbor in neighbors.iterrows()
]
)
done = False
else:
if sheet.face_df.loc[face, "num_sides"] > 3:
exchange(sheet, face, apoptosis_spec["geom"])
done = False
else:
remove(sheet, face, apoptosis_spec["geom"])
done = True
if not done:
manager.append(apoptosis, **apoptosis_spec)
We perform the events at each time points to run the simulation
[7]:
t = 0
stop=100
progress = widgets.IntProgress(min=0, max=stop)
progress.value = 0
display(progress)
history = History(sheet)
while manager.current and t < stop:
manager.execute(sheet)
t += 1
progress.value = t
res = solver.find_energy_min(sheet, geom, model, **min_settings)
history.record()
manager.update()
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/topology/sheet_topology.py:29: UserWarning: The epsilon argument is deprecated and will be removed in a future version. The length of the new edge should be set by `sheet.settings['threshold_length]*multiplier`
"The epsilon argument is deprecated and will be removed in a future version. "
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/topology/sheet_topology.py:29: UserWarning: The epsilon argument is deprecated and will be removed in a future version. The length of the new edge should be set by `sheet.settings['threshold_length]*multiplier`
"The epsilon argument is deprecated and will be removed in a future version. "
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/topology/sheet_topology.py:29: UserWarning: The epsilon argument is deprecated and will be removed in a future version. The length of the new edge should be set by `sheet.settings['threshold_length]*multiplier`
"The epsilon argument is deprecated and will be removed in a future version. "
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
3D view with ipyvolume¶
[8]:
color = sheet.vert_df['y']
fig, mesh = sheet_view(sheet, coords=['z', 'x', 'y'], edge={"color":color}, mode="3D")
fig
Provided you have ImageMagick installed, we can convert the output to a nice gif, like so:
[9]:
browse_history(history, edge={"color":lambda s : s.edge_df["length"]})
[10]:
create_gif(
history,
'single_apopto.gif',
num_frames=15,
draw_func=leg_joint_view
)
[11]:
Image("single_apopto.gif")
[11]:
<IPython.core.display.Image object>
[ ]: