Quasistatic vertex models¶
Quasitatic vertex models are based on the minimisation of a potential energy depending on the tissue geometry.
It is the original formalism defined by Honda et al., and in its most widely used form by Farhadifar et al..
tyssue provides a way to easily define such a potential and find the corresponding energy minimum through gradient descent.
[1]:
import sys
import pandas as pd
import numpy as np
import json
import matplotlib.pylab as plt
%matplotlib inline
from scipy import optimize
from tyssue import Sheet, config
from tyssue import SheetGeometry as geom
from tyssue.dynamics import effectors, model_factory
from tyssue.dynamics import SheetModel as model
from tyssue.solvers.quasistatic import QSSolver
from tyssue.draw import sheet_view
from tyssue.draw.plt_draw import plot_forces
from tyssue.io import hdf5
[2]:
h5store = 'data/small_hexagonal.hf5'
#h5store = 'data/before_apoptosis.hf5'
datasets = hdf5.load_datasets(h5store,
data_names=['face', 'vert', 'edge'])
specs = config.geometry.cylindrical_sheet()
sheet = Sheet('emin', datasets, specs)
sheet.sanitize(trim_borders=True, order_edges=True)
geom.update_all(sheet)
fig, ax = sheet_view(sheet, ['z', 'x'], mode='quick')
Effector classes¶
An effector designates a dynamical term in the epithelium governing equation. For quasistatic models, we need to provide a method to compute the energy associated with this effector, and the corresponding gradient.
For example, we can consider a line tension effector. The energy is \(E_t = \sum_{ij} \Lambda\ell_{ij}\) where the sum is over all the edges. For each half-edge, the gradient of \(E_t\) is defined by two terms, one for the gradient term associated with the half-edge \({ij}\) source, the over for it’s target. For the \(x\) coordinate:
Where \(\sum_k\) is are over all the edges which vertex \(i\) is a source, and \(\sum_m\) over all the edges of which vertex i is a target.
Here is the definition of the line tension effector:
class LineTension(AbstractEffector):
dimensions = units.line_tension
magnitude = "line_tension"
label = "Line tension"
element = "edge"
specs = {"edge": {"is_active": 1, "line_tension": 1e-2}}
spatial_ref = "mean_length", units.length
@staticmethod
def energy(eptm):
return eptm.edge_df.eval(
"line_tension" "* is_active" "* length / 2"
) # accounts for half edges
@staticmethod
def gradient(eptm):
grad_srce = -eptm.edge_df[eptm.ucoords] * to_nd(
eptm.edge_df.eval("line_tension * is_active/2"), len(eptm.coords)
)
grad_srce.columns = ["g" + u for u in eptm.coords]
grad_trgt = -grad_srce
return grad_srce, grad_trgt
For quasistatic methods, this effectors is agregated with others to define a model object. This object will have two methods compute_energy and compute_gradient that take an Epithelium object as single argument.
[3]:
model_specs = {
"edge": {
"line_tension": 1.,
},
"face": {
"contractility": 1.,
"vol_elasticity": 1.,
"prefered_vol": sheet.face_df["vol"].mean(),
},
}
sheet.update_specs(model_specs, reset=True)
Reseting column line_tension of the edge dataset with new specs
Reseting column contractility of the face dataset with new specs
Reseting column vol_elasticity of the face dataset with new specs
Reseting column prefered_vol of the face dataset with new specs
[4]:
Et, Ec, Ev = model.compute_energy(sheet, full_output=True)
energy = model.compute_energy(sheet, full_output=False)
print('Total energy: {}'.format(energy))
Total energy: 15221.226320514668
[5]:
grad_E = model.compute_gradient(sheet)
grad_E.head()
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
[5]:
| gx | gy | gz | |
|---|---|---|---|
| srce | |||
| 0 | -377.344701 | -1.563013e+02 | 697.693957 |
| 1 | -377.344701 | 1.563013e+02 | 697.693957 |
| 2 | -259.407529 | 1.074501e+02 | -622.859136 |
| 3 | 125.420612 | -3.979039e-13 | -617.293220 |
| 4 | -259.407529 | -1.074501e+02 | -622.859136 |
[6]:
model.labels
[6]:
['Line tension', 'Contractility', 'Volume elasticity']
[7]:
gradients = model.compute_gradient(sheet, components=True)
gradients = {label: (srce, trgt) for label, (srce, trgt)
in zip(model.labels, gradients)}
gradients['Line tension'][0].head()
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
[7]:
| gx | gy | gz | |
|---|---|---|---|
| edge | |||
| 0 | -0.00000 | -0.000000 | -0.500000 |
| 1 | -0.00000 | -0.000000 | 0.500000 |
| 2 | 0.08434 | 0.424007 | 0.251207 |
| 3 | -0.08434 | 0.424007 | -0.251207 |
| 4 | 0.08434 | -0.424007 | 0.251207 |
[8]:
%pdb
Automatic pdb calling has been turned ON
[9]:
solver = QSSolver()
scale = 0.2
fig, ax = plot_forces(sheet, geom, model, ['z', 'x'], scale)
fig.set_size_inches(10, 12)
for n, (vx, vy, vz) in sheet.vert_df[sheet.coords].iterrows():
shift = 0.6 * np.sign(vy)
ax.text(vz+shift-0.3, vx, str(n))
app_grad_specs = config.draw.sheet_spec()['grad']
app_grad_specs.update({'color':'r'})
def draw_approx_force(ax=None):
fig, ax = plot_forces(sheet, geom, model,
['z', 'x'], scaling=scale, ax=ax,
approx_grad=solver.approx_grad, **{'grad':app_grad_specs})
fig.set_size_inches(10, 12)
return fig, ax
## Uncomment bellow to recompute
fig, ax = draw_approx_force(ax=ax)
#fig
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
http://scipy.github.io/devdocs/generated/scipy.optimize.check_grad.html#scipy.optimize.check_grad
[10]:
grad_err = solver.check_grad(sheet, geom, model)
grad_err /= sheet.vert_df.size
print("Error on the gradient (non-dim, per vertex): {:.3e}".format(grad_err))
/home/docs/checkouts/readthedocs.org/user_builds/tyssue/conda/stable/lib/python3.7/site-packages/tyssue/utils/utils.py:35: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
df_nd = df[:, None]#np.asarray(df).repeat(ndim).reshape((df.size, ndim))
Error on the gradient (non-dim, per vertex): 1.970e-02
[11]:
options ={'disp':False,
'ftol':1e-5,
'gtol':1e-5}
res = solver.find_energy_min(sheet, geom, model, options=options)
print(res['success'])
True
[12]:
res['message']
[12]:
b'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'
[13]:
res['fun']/sheet.face_df.is_alive.sum()
[13]:
240.67238964483766
[14]:
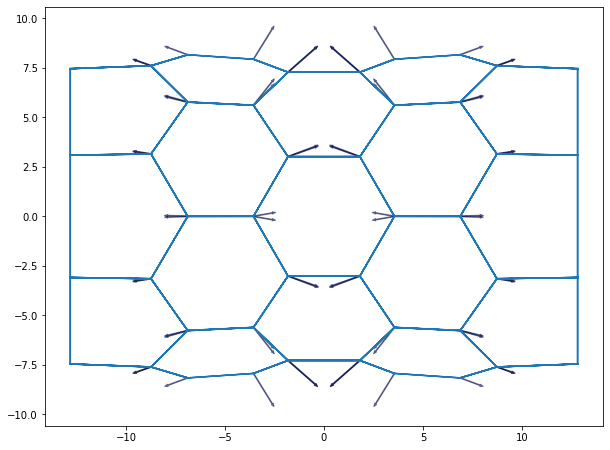
fig, ax = plot_forces(sheet, geom, model, ['z', 'y'], 1)
fig.set_size_inches(10, 12)
[ ]: