Cell Division#
On a 2D mesh#
import pandas as pd
import numpy as np
import json
import matplotlib.pylab as plt
import ipyvolume as ipv
%matplotlib inline
from tyssue import Sheet, config
from tyssue.geometry.planar_geometry import PlanarGeometry as geom
from tyssue.solvers.quasistatic import QSSolver
from tyssue.dynamics.planar_vertex_model import PlanarModel as model
from tyssue.draw import sheet_view
from tyssue.stores import load_datasets
from tyssue.topology.sheet_topology import remove_face, cell_division
/tmp/ipykernel_3605/1651668793.py:1: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import pandas as pd
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[1], line 5
3 import json
4 import matplotlib.pylab as plt
----> 5 import ipyvolume as ipv
6 get_ipython().run_line_magic('matplotlib', 'inline')
8 from tyssue import Sheet, config
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/__init__.py:8
6 from ipyvolume import datasets # noqa: F401
7 from ipyvolume import embed # noqa: F401
----> 8 from ipyvolume.widgets import * # noqa: F401, F403
9 from ipyvolume.transferfunction import * # noqa: F401, F403
10 from ipyvolume.pylab import * # noqa: F401, F403
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/widgets.py:23
21 import ipyvolume._version
22 from ipyvolume.traittypes import Image
---> 23 from ipyvolume.serialize import (
24 array_cube_tile_serialization,
25 array_serialization,
26 array_sequence_serialization,
27 color_serialization,
28 texture_serialization,
29 )
30 from ipyvolume.transferfunction import TransferFunction
31 from ipyvolume.utils import debounced, grid_slice, reduce_size
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/serialize.py:17
15 import ipywidgets
16 import ipywebrtc
---> 17 from ipython_genutils.py3compat import string_types
19 from ipyvolume import utils
22 logger = logging.getLogger("ipyvolume")
ModuleNotFoundError: No module named 'ipython_genutils'
solver = QSSolver()
sheet = Sheet.planar_sheet_2d('division', 6, 6, 1, 1)
sheet.sanitize(trim_borders=True, order_edges=True)
geom.update_all(sheet)
sheet.get_opposite()
# ## Set up the model
nondim_specs = config.dynamics.quasistatic_plane_spec()
dim_model_specs = model.dimensionalize(nondim_specs)
sheet.update_specs(dim_model_specs, reset=True)
print("Number of cells: {}\n"
" edges: {}\n"
" vertices: {}\n".format(sheet.Nf, sheet.Ne, sheet.Nv))
# ## Minimize energy
res = solver.find_energy_min(sheet, geom, model)
# ## View the result
draw_specs = config.draw.sheet_spec()
draw_specs['vert']['visible'] = False
draw_specs['edge']['head_width'] = 0.1
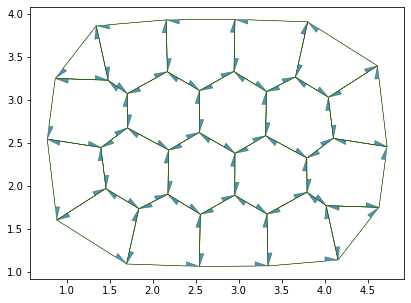
fig, ax = sheet_view(sheet, **draw_specs)
fig.set_size_inches(12, 5)
Reseting column is_alive of the face dataset with new specs
Reseting column ux of the edge dataset with new specs
Reseting column uy of the edge dataset with new specs
Reseting column is_active of the vert dataset with new specs
Number of cells: 20
edges: 100
vertices: 38
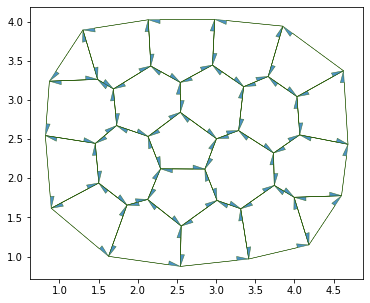
daughter = cell_division(sheet, 7, geom, angle=np.pi/2)
res = solver.find_energy_min(sheet, geom, model)
print(res['success'])
fig, ax = sheet_view(sheet, **draw_specs)
fig.set_size_inches(12, 5)
True
Division in a 3D single layer epithelium#
from tyssue.io.hdf5 import save_datasets, load_datasets
# redefine cell_division from monolayer related topology module
from tyssue.topology.monolayer_topology import cell_division
from tyssue.core.monolayer import Monolayer
from tyssue.geometry.bulk_geometry import ClosedMonolayerGeometry as monolayer_geom
from tyssue.dynamics.bulk_model import ClosedMonolayerModel
from tyssue.draw import highlight_cells
datasets = load_datasets('data/small_ellipsoid.hf5',
data_names=['vert', 'edge',
'face', 'cell'])
specs = config.geometry.bulk_spec()
monolayer = Monolayer('ell', datasets, specs)
monolayer_geom.update_all(monolayer)
specs = {
"edge": {
"line_tension": 0.0,
},
"face": {
"contractility": 0.01,
},
"cell": {
"prefered_vol": monolayer.cell_df['vol'].median(),
"vol_elasticity": 0.1,
"prefered_area": monolayer.cell_df['area'].median(),
"area_elasticity": 0.1,
},
"settings": {
'lumen_prefered_vol': monolayer.settings['lumen_vol'],
'lumen_vol_elasticity': 1e-2
}
}
monolayer.update_specs(specs, reset=True)
res = solver.find_energy_min(monolayer, monolayer_geom, ClosedMonolayerModel)
Reseting column line_tension of the edge dataset with new specs
Reseting column contractility of the face dataset with new specs
Reseting column prefered_vol of the cell dataset with new specs
Reseting column vol_elasticity of the cell dataset with new specs
mother = 8
daughter = cell_division(monolayer, mother,
orientation='vertical')
monolayer.validate()
True
rho = np.linalg.norm(monolayer.vert_df[monolayer.coords], axis=1)
draw_specs['edge']['color'] = rho
draw_specs['face']['visible'] = True
ipv.clear()
highlight_cells(monolayer, mother, reset_visible=True)
fig, mesh = sheet_view(monolayer, mode="3D",
coords=['z', 'x', 'y'], **draw_specs)
fig
mother = 18
daughter = cell_division(monolayer, mother,
orientation='horizontal')
monolayer.validate()
True
rho = np.linalg.norm(monolayer.vert_df[monolayer.coords], axis=1)
draw_specs['edge']['color'] = rho
draw_specs['face']['visible'] = True
ipv.clear()
highlight_cells(monolayer, mother)
fig, mesh = sheet_view(monolayer, mode="3D",
coords=['z', 'x', 'y'], **draw_specs)
fig
Energy minimisation of the monolayer after division#
monolayer.cell_df.loc[[mother, daughter], 'prefered_area'] /= 2
monolayer.cell_df.loc[[mother, daughter], 'prefered_vol'] /= 3
monolayer.settings['lumen_prefered_vol'] = monolayer.settings['lumen_vol']
monolayer.settings['lumen_vol_elasticity'] = 1e-2
res = solver.find_energy_min(monolayer, monolayer_geom, ClosedMonolayerModel)
print(res['message'])
b'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
rho = np.linalg.norm(monolayer.vert_df[monolayer.coords], axis=1)
draw_specs['edge']['color'] = rho
ipv.clear()
highlight_cells(monolayer, mother)
fig, mesh = sheet_view(monolayer, mode="3D",
coords=['z', 'x', 'y'], **draw_specs)
fig
from tyssue.generation import three_faces_sheet, extrude
from tyssue.geometry.bulk_geometry import MonoLayerGeometry
datasets_2d, _ = three_faces_sheet(zaxis=True)
datasets = extrude(datasets_2d, method='translation')
eptm = Monolayer('test_volume', datasets,
config.geometry.bulk_spec(),
coords=['x','y','z'])
## We need to add noise to our position to avoid
## Ill defined angles in this artificial case
eptm.vert_df[eptm.coords] += np.random.normal(
scale=1e-6,
size=eptm.vert_df[eptm.coords].shape)
MonoLayerGeometry.update_all(eptm)
for orientation in ['vertical', 'horizontal']:
print(orientation)
daughter = cell_division(eptm, 1, orientation=orientation)
eptm.reset_topo()
eptm.reset_index()
MonoLayerGeometry.update_all(eptm)
print(f'Valid division for {orientation}:', end="\t")
print(eptm.validate())
vertical
Valid division for vertical: True
horizontal
Valid division for horizontal: True
ipv.clear()
highlight_cells(eptm, daughter)
fig, mesh = sheet_view(eptm, mode="3D")
fig
