Energy minimization for a 2.5D sheet in 3D#
import sys
import pandas as pd
import numpy as np
import json
import matplotlib.pylab as plt
%matplotlib inline
from scipy import optimize
from tyssue import Sheet
from tyssue import SheetGeometry as geom
from tyssue.dynamics.sheet_vertex_model import SheetModel as model
from tyssue.solvers.quasistatic import QSSolver
from tyssue import config
from tyssue.solvers.isotropic_solver import bruteforce_isotropic_relax
from tyssue.draw.plt_draw import sheet_view
import tyssue.draw.plt_draw as draw
from tyssue.io import hdf5
/tmp/ipykernel_3741/3375153074.py:2: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import pandas as pd
collision solver could not be imported You may need to install CGAL and re-install tyssue
This module needs ipyvolume to work.
You can install it with:
$ conda install -c conda-forge ipyvolume
h5store = 'data/small_hexagonal.hf5'
datasets = hdf5.load_datasets(h5store,
data_names=['face', 'vert', 'edge'])
specs = config.geometry.cylindrical_sheet()
sheet = Sheet('emin', datasets, specs)
sheet.sanitize(trim_borders=True, order_edges=True)
geom.update_all(sheet)
sheet.vert_df.describe().head(3)
| y | is_active | z | x | rho | old_idx | basal_shift | height | radial_tension | |
|---|---|---|---|---|---|---|---|---|---|
| count | 8.000000e+01 | 80.000000 | 8.000000e+01 | 8.000000e+01 | 8.000000e+01 | 80.000000 | 8.000000e+01 | 8.000000e+01 | 80.0 |
| mean | 1.110223e-15 | 0.800000 | 8.881784e-17 | -2.620126e-15 | 8.071634e+00 | 103.425000 | 2.216705e+00 | 5.854929e+00 | 0.0 |
| std | 5.743517e+00 | 0.402524 | 8.027290e+00 | 5.743517e+00 | 4.968335e-15 | 27.759261 | 4.468911e-16 | 5.000389e-15 | 0.0 |
sheet.face_df.describe().head(3)
| area | z | y | is_alive | x | perimeter | old_idx | num_sides | vol | prefered_area | prefered_vol | contractility | vol_elasticity | prefered_height | basal_shift | basal_height | height | rho | id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 40.000000 | 40.000000 | 4.000000e+01 | 40.0 | 4.000000e+01 | 40.000000 | 40.000000 | 40.000000 | 40.000000 | 40.0 | 40.0 | 40.000000 | 40.0 | 40.0 | 40.000000 | 40.000000 | 4.000000e+01 | 4.000000e+01 | 40.0 |
| mean | 31.948614 | 0.000000 | 1.176836e-15 | 1.0 | -2.653433e-15 | 21.447238 | 27.500000 | 5.600000 | 187.056872 | 12.0 | 288.0 | 76.076404 | 1.0 | 24.0 | 2.500252 | -3.246161 | 5.854929e+00 | 8.071634e+00 | 0.0 |
| std | 2.890942 | 7.446702 | 5.463498e+00 | 0.0 | 5.463498e+00 | 0.549335 | 16.306362 | 0.496139 | 16.926260 | 0.0 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.000000 | 0.000000 | 2.491937e-15 | 2.257710e-15 | 0.0 |
sheet.edge_df.describe().head(3)
| dz | length | dx | dy | srce | trgt | face | old_jv0 | old_jv1 | old_cell | ... | sz | tx | ty | tz | fx | fy | fz | rx | ry | rz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 2.240000e+02 | 224.000000 | 2.240000e+02 | 2.240000e+02 | 224.000000 | 224.000000 | 224.000000 | 224.000000 | 224.000000 | 224.000000 | ... | 224.000000 | 2.240000e+02 | 2.240000e+02 | 224.000000 | 2.240000e+02 | 2.240000e+02 | 224.000000 | 2.240000e+02 | 2.240000e+02 | 2.240000e+02 |
| mean | 3.965082e-18 | 3.829864 | -1.982541e-17 | -7.930164e-18 | 39.500000 | 39.500000 | 19.500000 | 99.348214 | 106.031250 | 27.500000 | ... | 0.000000 | -2.727977e-15 | 1.268826e-15 | 0.000000 | -2.474211e-15 | 1.015061e-15 | 0.000000 | 3.965082e-17 | -2.577303e-17 | 2.973812e-17 |
| std | 2.593865e+00 | 0.652700 | 2.053231e+00 | 2.053231e+00 | 23.111351 | 23.111351 | 11.563046 | 27.073959 | 27.627631 | 16.132856 | ... | 7.530508 | 5.720290e+00 | 5.720290e+00 | 7.530508 | 5.409339e+00 | 5.409339e+00 | 7.117109 | 1.860313e+00 | 1.860313e+00 | 2.460756e+00 |
3 rows × 33 columns
Non dimensionalisation and isotropic model#
For our 2D1/2 model, for a tissue energy is expressed as: $\( E = N_f\frac{K}{2}(V - V_0)^2 + N_f \frac{\Gamma}{2}L^2 + 3N_f\Lambda \ell \)\( In the case of a regular, infinite hexagonal lattice, There 6 edges per cell, each shared bteween 2 cells, hence \)Ne = 3N_f$
We can write this equation as a dimension-less one by dividing it by \(N_f K V_0^2\). We define the dimension-less values: \(\bar\Gamma = \Gamma/K V_0^{4/3}\) and \(\bar\Lambda = \Lambda/K V_0^{5/3}\)
We define the scaling factor \(\delta\) such that \(V/V_0 = \delta^3\). On a regular hexagonal grid, the perimeter \(L=6\ell\) and the area is A=\(3\sqrt{3}/2\,\ell^2\). The volume is \(V = Ah\). As the (pseudo-)height is arbitrary, we can chose its prefered value. We set \(h_0 = V_0^{1/3}\).
We thus have \(A_0 = V_0^{2/3}\), \(A = \delta^2 V_0^{2/3}\) and \(\ell = \delta (\frac{2}{3\sqrt{3}})^{1/2} V_0^{1/3}\). We define the constant \(\mu = 2^{3/2}3^{1/4}\) to simplify further the equation:
The root of this polynomial corresponds to the lowest energy possible for this set of paramters.
### This analytic model is implemented bellow
It is now a bit too specific to warrant a module in tyssue, and was thus removed
"""
Isotropic energy model from Farhadifar et al. 2007 article
"""
import numpy as np
mu = 2 ** 1.5 * 3 ** 0.25
def elasticity(delta):
return (delta ** 3 - 1) ** 2 / 2.0
def contractility(delta, gamma):
return gamma * mu ** 2 * delta ** 2 / 2.0
def tension(delta, lbda):
return lbda * mu * delta / 2.0
def isotropic_energies(sheet, model, geom, deltas, nondim_specs):
# bck_face_shift = sheet.face_df['basal_shift']
# bck_vert_shift = sheet.vert_df['basal_shift']
# ## Faces only area and height
V_0 = sheet.specs["face"]["prefered_vol"]
vol_avg = sheet.face_df[sheet.face_df["is_alive"] == 1].vol.mean()
rho_avg = sheet.vert_df.rho.mean()
# ## Set height and volume to height0 and V0
delta_0 = (V_0 / vol_avg) ** (1 / 3)
geom.scale(sheet, delta_0, sheet.coords)
h_0 = V_0 ** (1 / 3)
sheet.face_df["basal_shift"] = rho_avg * delta_0 - h_0
sheet.vert_df["basal_shift"] = rho_avg * delta_0 - h_0
geom.update_all(sheet)
energies = np.zeros((deltas.size, 3))
for n, delta in enumerate(deltas):
geom.scale(sheet, delta, sheet.coords + ["basal_shift"])
geom.update_all(sheet)
Et, Ec, Ev = model.compute_energy(sheet, full_output=True)
energies[n, :] = [Et.sum(), Ec.sum(), Ev.sum()]
geom.scale(sheet, 1 / delta, sheet.coords + ["basal_shift"])
geom.update_all(sheet)
energies /= sheet.face_df["is_alive"].sum()
isotropic_relax(sheet, nondim_specs)
return energies
def isotropic_relax(sheet, nondim_specs, geom=geom):
"""Deforms the sheet so that the faces pseudo-volume is at their
isotropic optimum (on average)
The specified model specs is assumed to be non-dimentional
"""
vol0 = sheet.face_df["prefered_vol"].mean()
h_0 = vol0 ** (1 / 3)
live_faces = sheet.face_df[sheet.face_df.is_alive == 1]
vol_avg = live_faces.vol.mean()
rho_avg = sheet.vert_df.rho.mean()
# ## Set height and volume to height0 and vol0
delta = (vol0 / vol_avg) ** (1 / 3)
geom.scale(sheet, delta, coords=sheet.coords)
sheet.face_df["basal_shift"] = rho_avg * delta - h_0
sheet.vert_df["basal_shift"] = rho_avg * delta - h_0
geom.update_all(sheet)
# ## Optimal value for delta
delta_o = find_grad_roots(nondim_specs)
if not np.isfinite(delta_o):
raise ValueError("invalid parameters values")
sheet.delta_o = delta_o
# ## Scaling
geom.scale(sheet, delta_o, coords=sheet.coords + ["basal_shift"])
geom.update_all(sheet)
def isotropic_energy(delta, mod_specs):
"""
Computes the theoritical energy per face for the given
parameters.
"""
lbda = mod_specs["edge"]["line_tension"]
gamma = mod_specs["face"]["contractility"]
elasticity_ = (delta ** 3 - 1) ** 2 / 2.0
contractility_ = gamma * mu ** 2 * delta ** 2 / 2.0
tension_ = lbda * mu * delta / 2.0
energy = elasticity_ + contractility_ + tension_
return energy
def isotropic_grad_poly(mod_specs):
lbda = mod_specs["edge"]["line_tension"]
gamma = mod_specs["face"]["contractility"]
grad_poly = [3, 0, 0, -3, mu ** 2 * gamma, mu * lbda / 2.0]
return grad_poly
def isotropic_grad(mod_specs, delta):
grad_poly = isotropic_grad_poly(mod_specs)
return np.polyval(grad_poly, delta)
def find_grad_roots(mod_specs):
poly = isotropic_grad_poly(mod_specs)
roots = np.roots(poly)
good_roots = np.real([r for r in roots if np.abs(r) == r])
np.sort(good_roots)
if len(good_roots) == 1:
return good_roots
elif len(good_roots) > 1:
return good_roots[0]
else:
return np.nan
nondim_specs = config.dynamics.quasistatic_sheet_spec()
dim_model_specs = model.dimensionalize(nondim_specs)
sheet.update_specs(dim_model_specs, reset=True)
sheet.edge_df.is_active = (sheet.upcast_face(sheet.face_df.is_alive)
& sheet.upcast_srce(sheet.vert_df.is_active))
res = bruteforce_isotropic_relax(sheet, geom, model)
Et, Ec, Ev = model.compute_energy(sheet, full_output=True)
energy = model.compute_energy(sheet, full_output=False)
print('Total energy: {}'.format(energy))
Total energy: 18.662021831602555
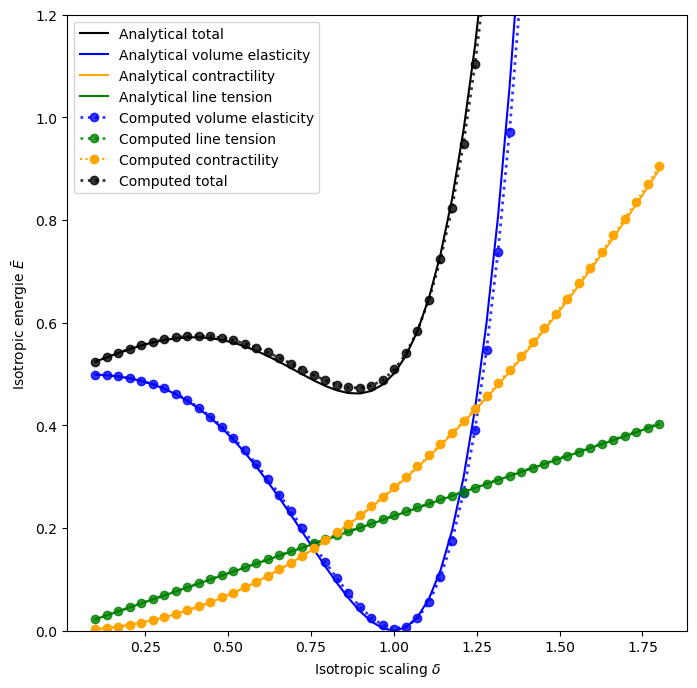
def plot_analytical_to_numeric_comp(sheet, model, geom, nondim_specs):
fig, ax = plt.subplots(figsize=(8, 8))
deltas = np.linspace(0.1, 1.8, 50)
lbda = nondim_specs["edge"]["line_tension"]
gamma = nondim_specs["face"]["contractility"]
ax.plot(
deltas,
isotropic_energy(deltas, nondim_specs),
"k-",
label="Analytical total",
)
try:
ax.plot(sheet.delta_o, isotropic_energy(sheet.delta_o, nondim_specs), "ro")
except:
pass
ax.plot(deltas, elasticity(deltas), "b-", label="Analytical volume elasticity")
ax.plot(
deltas,
contractility(deltas, gamma),
color="orange",
ls="-",
label="Analytical contractility",
)
ax.plot(deltas, tension(deltas, lbda), "g-", label="Analytical line tension")
ax.set_xlabel(r"Isotropic scaling $\delta$")
ax.set_ylabel(r"Isotropic energie $\bar E$")
energies = isotropic_energies(sheet, model, geom, deltas, nondim_specs)
# energies = energies / norm
ax.plot(
deltas,
energies[:, 2],
"bo:",
lw=2,
alpha=0.8,
label="Computed volume elasticity",
)
ax.plot(
deltas, energies[:, 0], "go:", lw=2, alpha=0.8, label="Computed line tension"
)
ax.plot(
deltas,
energies[:, 1],
ls=":",
marker="o",
color="orange",
label="Computed contractility",
)
ax.plot(
deltas, energies.sum(axis=1), "ko:", lw=2, alpha=0.8, label="Computed total"
)
ax.legend(loc="upper left", fontsize=10)
ax.set_ylim(0, 1.2)
print(sheet.delta_o, deltas[energies.sum(axis=1).argmin()])
return fig, ax
fig, ax = plot_analytical_to_numeric_comp(sheet, model, geom, nondim_specs)
0.8865926873873902 0.8979591836734694
model.labels
['Line tension', 'Contractility', 'Volume elasticity']
gradients = model.compute_gradient(sheet, components=True)
gradients = {label: (srce, trgt) for label, (srce, trgt)
in zip(model.labels, gradients)}
gradients['Line tension'][0].head()
| gx | gy | gz | |
|---|---|---|---|
| edge | |||
| 0 | -0.000000 | -0.000000 | -668.132926 |
| 1 | -0.000000 | -0.000000 | 668.132926 |
| 2 | 112.700882 | 566.585593 | 335.679736 |
| 3 | -112.700882 | 566.585593 | -335.679736 |
| 4 | 112.700882 | -566.585593 | 335.679736 |
grad_i = model.compute_gradient(sheet, components=False)
grad_i.head()
| gx | gy | gz | |
|---|---|---|---|
| srce | |||
| 0 | -0.008983 | -3.720720e-03 | -0.001010 |
| 1 | -0.008983 | 3.720720e-03 | -0.001010 |
| 2 | -0.018412 | 7.626655e-03 | -0.009825 |
| 3 | -0.013376 | -7.264422e-17 | -0.008121 |
| 4 | -0.018412 | -7.626655e-03 | -0.009825 |
geom.scale(sheet, 1.2, sheet.coords)
geom.update_all(sheet)
scale = 5
fig, ax = draw.plot_forces(sheet, geom, model, ['z', 'x'], scale)
fig.set_size_inches(10, 12)
for n, (vx, vy, vz) in sheet.vert_df[sheet.coords].iterrows():
shift = 0.6 * np.sign(vy)
ax.text(vz+shift-0.3, vx, str(n))
app_grad_specs = config.draw.sheet_spec()['grad']
app_grad_specs.update({'color':'r'})
solver = QSSolver()
sheet.topo_changed = False
def draw_approx_force(ax=None):
fig, ax = draw.plot_forces(sheet, geom, model,
['z', 'x'], scaling=scale, ax=ax,
approx_grad=solver.approx_grad, **{'grad':app_grad_specs})
fig.set_size_inches(10, 12)
return fig, ax
## Uncomment bellow to recompute
fig, ax = draw_approx_force(ax=ax)
#fig
http://scipy.github.io/devdocs/generated/scipy.optimize.check_grad.html#scipy.optimize.check_grad
grad_err = solver.check_grad(sheet, geom, model)
grad_err /= sheet.Nv
print("Error on the gradient (non-dim, per vertex): {:.3e}".format(grad_err))
Error on the gradient (non-dim, per vertex): 1.688e-05
settings = {
'options': {'disp':False,
'ftol':1e-5,
'gtol':1e-5},
}
res = solver.find_energy_min(sheet, geom, model, **settings)
print(res['success'])
True
res['message']
'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'
res['fun']/sheet.face_df.is_alive.sum()
0.43145767586018635
fig, ax = draw.plot_forces(sheet, geom, model, ['z', 'y'], 10)
fig.set_size_inches(10, 12)