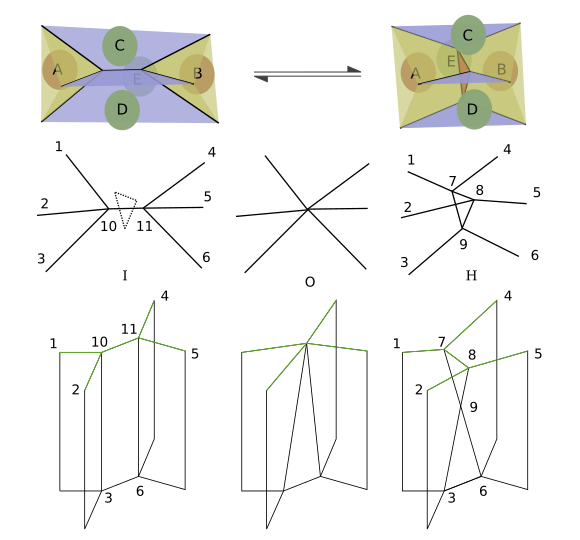
Reversible Network Reconnection Model#
Attempt at implementing the RNR model as described in Okuda et al. 2012:
Reversible network reconnection model for simulating large deformation in dynamic tissue morphogenesis, Satoru Okuda, Yasuhiro Inoue, Mototsugu Eiraku, Yoshiki Sasai and Taiji Adachi Biomech Model Mechanobiol (2013) 12:627–644 DOI 10.1007/s10237-012-0430-7
The authors give 4 conditions which we detail and implement bellow.
Condition 1 - Center of a face#
The center of a face is defined by the average position of the face’s edges midpoints, weighted by their lengths.
import numpy as np, pandas as pd
import ipyvolume as ipv
import matplotlib.pyplot as plt
%matplotlib inline
from tyssue import Epithelium
from tyssue import BulkGeometry, RNRGeometry
from tyssue import Sheet
from tyssue.config.geometry import bulk_spec
from tyssue.generation import three_faces_sheet
from tyssue.generation import extrude
from tyssue.draw import sheet_view, highlight_cells
from tyssue.topology.bulk_topology import IH_transition, HI_transition
draw_spec = {'face': {'visible': True}}
sheet = Sheet.planar_sheet_3d('sheet', 5, 5, 1, 1)
sheet.sanitize()
datasets = extrude(sheet.datasets, method='translation')
eptm = Epithelium('20faces_3D', datasets, bulk_spec())
eptm.vert_df[eptm.coords] += np.random.normal(scale=1e-2, size=(eptm.Nv, 3))
RNRGeometry.update_all(eptm)
RNRGeometry.center(eptm)
RNRGeometry.update_all(eptm)
eptm.settings['threshold_length'] = 1e-4
print(eptm.Nf, eptm.Ne, eptm.Nv)
draw_spec['face']['color'] = eptm.face_df.area
draw_spec['face']['color_range'] = (0, 1)
ipv.clear()
fig, meshes = sheet_view(eptm, mode='3D', **draw_spec)
fig
/tmp/ipykernel_3653/2203072209.py:1: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import numpy as np, pandas as pd
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[1], line 2
1 import numpy as np, pandas as pd
----> 2 import ipyvolume as ipv
3 import matplotlib.pyplot as plt
4 get_ipython().run_line_magic('matplotlib', 'inline')
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/__init__.py:8
6 from ipyvolume import datasets # noqa: F401
7 from ipyvolume import embed # noqa: F401
----> 8 from ipyvolume.widgets import * # noqa: F401, F403
9 from ipyvolume.transferfunction import * # noqa: F401, F403
10 from ipyvolume.pylab import * # noqa: F401, F403
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/widgets.py:23
21 import ipyvolume._version
22 from ipyvolume.traittypes import Image
---> 23 from ipyvolume.serialize import (
24 array_cube_tile_serialization,
25 array_serialization,
26 array_sequence_serialization,
27 color_serialization,
28 texture_serialization,
29 )
30 from ipyvolume.transferfunction import TransferFunction
31 from ipyvolume.utils import debounced, grid_slice, reduce_size
File ~/checkouts/readthedocs.org/user_builds/tyssue/conda/latest/lib/python3.12/site-packages/ipyvolume/serialize.py:17
15 import ipywidgets
16 import ipywebrtc
---> 17 from ipython_genutils.py3compat import string_types
19 from ipyvolume import utils
22 logger = logging.getLogger("ipyvolume")
ModuleNotFoundError: No module named 'ipython_genutils'
I→H transition#
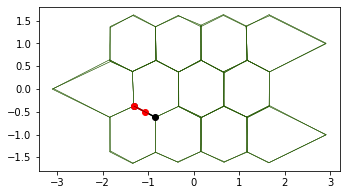
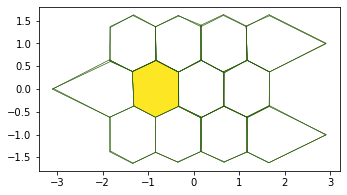
This produces a shape similar to the one studied by LM Escudero and collegues in their article.
e_1011 = 26
eptm.settings['threshold_length'] = 0.2
srce, trgt, face, cell = eptm.edge_df.loc[e_1011,
['srce', 'trgt',
'face', 'cell']]
IH_transition(eptm, e_1011)
RNRGeometry.update_all(eptm)
highlight_cells(eptm, cell, reset_visible=True)
draw_spec['face']['color'] = eptm.face_df.area
ipv.clear()
fig, meshes = sheet_view(eptm, mode='3D', **draw_spec)
fig
cell 0 is already closed
cell 5 is already closed
H → I transition#
fa = 93
HI_transition(eptm, fa)
RNRGeometry.update_all(eptm)
highlight_cells(eptm, cell, reset_visible=True)
ipv.clear()
draw_spec['face']['color'] = eptm.face_df.area
draw_spec['face']['visible'] = True
eptm.face_df.visible = True
#fig, meshes = sheet_view(eptm, mode='3D', **draw_spec)
#fig
cell 4 is already closed
cell 0 is already closed
cell 5 is already closed
cell 1 is already closed
eptm.edge_df[eptm.edge_df['sub_vol'] <= 1e-10][["srce", "trgt", "face", "cell", "length", "sub_area", "sx", "sy", "sz", "tx", "ty", "tz", "fx", "fy", "fz"]]
| srce | trgt | face | cell | length | sub_area | sx | sy | sz | tx | ty | tz | fx | fy | fz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| edge | |||||||||||||||
| 241 | 39 | 51 | 50 | 5 | 0.534805 | 7.372575e-18 | -1.312808 | -0.374665 | -0.493168 | -0.838700 | -0.621678 | -0.508126 | -1.075754 | -0.498172 | -0.500647 |
| 244 | 51 | 39 | 51 | 0 | 0.534805 | 7.564593e-18 | -0.838700 | -0.621678 | -0.508126 | -1.312808 | -0.374665 | -0.493168 | -1.075754 | -0.498172 | -0.500647 |
fig, ax = sheet_view(eptm, coords=["x", "y"])
ax.plot(eptm.edge_df.loc[241, ["sx", "tx"]], eptm.edge_df.loc[241, ["sy", "ty"]], "ko-")
ax.plot(eptm.edge_df.loc[241, ["fx", "sx"]], eptm.edge_df.loc[241, ["fy", "sy"]], "ro-")
for e, data in eptm.edge_df[eptm.edge_df.face.isin([50, 51])].iterrows():
ax.plot(data[["sx", "tx"]], data[["sy", "ty"]], "k-",lw=0.2)
RNRGeometry.update_areas(eptm)
highlight_cells(eptm, cell, reset_visible=False)
ipv.clear()
draw_spec['face']['color'] = eptm.face_df.area
eptm.face_df.visible = True
#fig, meshes = sheet_view(eptm, mode='3D', **draw_spec)
#fig
Testing for I→H / H →I transition triggers#
highlight_cells(eptm, cell, reset_visible=True)
draw_spec['face']['color'] = eptm.face_df.area
fig, ax = sheet_view(eptm, coords = ["x", "z"], **draw_spec)
fig, ax = sheet_view(eptm, coords = ["x", "y"], **draw_spec)

eptm.settings['threshold_length'] = 1e-3
#eptm.settings['threshold_length'] = eptm.edge_df.length.min()+1e-3
def find_rearangements(eptm):
l_th = eptm.settings['threshold_length']
up_num_sides = eptm.upcast_face(eptm.face_df['num_sides'])
shorts = eptm.edge_df[eptm.edge_df['length'] < l_th]
non_triangular = up_num_sides[up_num_sides > 4 ].index
edges_IH = set(shorts.index).intersection(non_triangular)
max_f_length = shorts.groupby('face')['length'].apply(max)
short_faces = max_f_length[max_f_length < l_th].index
three_faces = eptm.face_df[eptm.face_df['num_sides'] == 3].index
faces_HI = set(three_faces).intersection(short_faces)
return edges_IH, faces_HI
find_rearangements(eptm)
(set(), set())
Condition 3#
This condition is satisfied if eptm.settings['threshold_length']
is well defined, i.e, small with respect to the unit length or the
average edge length.
Condition 4#
(i) Two edges never share two vertices simultaneously.
In our half-edge architecture, this is not as straight forward. But I think the condition can be reformulated as: Two edges from the same face never share two edges simultanously. I don’t know how to demonstrate this, but I think these are equivalent (to get two edges together, you need to “squeeze” one face between those two.
(ii) Two polygonal faces never share two or more edges simultaneously.
from tyssue.topology.base_topology import condition_4i, condition_4ii
condition_4i(eptm), condition_4ii(eptm)
(Int64Index([], dtype='int64', name='face'),
array([], shape=(0, 2), dtype=int64))
eptm.settings['threshold_length'] = 0.5
IH_transition(eptm, eptm.edge_df.index[-1])
RNRGeometry.update_all(eptm)
draw_spec['face']['color'] = eptm.face_df.area
#ipv.clear()
#fig, meshes = sheet_view(eptm, mode='3D', **draw_spec)
#fig
cell 0 is already closed
cell 5 is already closed
/home/guillaume/Dev/tyssue/tyssue/geometry/bulk_geometry.py:107: RuntimeWarning: invalid value encountered in divide
weighted_pos.values / eptm.face_df["perimeter"].values[:, np.newaxis]

